Analysis of the Bioactive Compounds from Carica papaya in the Management of Psoriasis using Computational Techniques
Keywords:
Psoriasis, Carica Papaya, Molecular docking, Anti-inflammatory, Skin disorderAbstract
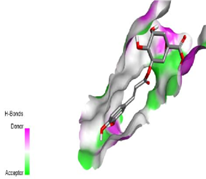
Psoriasis is a persistent and mysterious autoimmune skin condition that affects 2-3% of the world’s population. Currently, topical therapies, light therapy, and systemic drugs are the three main forms of treatment used to lessen inflammation and skin irritation/itching. However, all these treatments are only used to manage the disease each time it surfaces. Therefore, the main target of this work is to search for a safer and more effective remedy for psoriasis from the reservoir of phytochemicals present in Carica papaya via in silico studies due to its anti-psoriatic and anti-inflammatory properties. Reported phytochemicals isolated from Carica papaya were subjected to computational simulations using the PyRx docking tool and were docked against Janus Kinase 1 (JAK1) and Tumor necrosis factor \aplha (TNF\aplha) target receptors. The results obtained were visualized using PyMol, and Biovia 2019. Analysis of the results identified both Chlorogenic acid and Coumaroylquinic-acid with docking scores (-8.6 kcal/mol and -7.9 kcal/mol) respectively as potential inhibitors for the JAK1 receptor. The identified compounds also possessed excellent ADMET, drug-likeness, bioactivity, and activity spectra for substances (PASS) prediction properties. Their binding mode and the molecular interactions with the targets also affirmed their potency. In comparison with the standards (Methotrexate and Cyclosporine), Chlorogenic acid and Coumaroylquinic-acid have better ADMET properties, binding affinities, drug-likeness, PASS properties, bioactivities, oral bioavailability, binding mechanism, and interactions with the active site of the target receptor and are hereby recommended for further analysis towards the development of a new therapeutic agent for psoriasis treatment and management.
Published
How to Cite
Issue
Section
Copyright (c) 2023 Misbaudeen Abdul-Hammed, Ibrahim Olaide Adedotun, Tolulope Irapada Afolabi Afolabi, Ubeydat Temitope Ismail, Praise Toluwalase Akande, Balqees Funmilayo Issa

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Similar Articles
- B. Bako, E. E. Etim, J. P. Shinggu, S. S. Humphrey, L. J. Moses, M. E. Khan, Quantum chemical calculations of lupeol (C30H50O) isolated from the ethyl acetate leaf extracts of Justicia Secunda , Journal of the Nigerian Society of Physical Sciences: Volume 6, Issue 3, August 2024
- Nassima Bou-ydia, Ben-issa El miraoui, Latifa Laallam, Ahmed Jouaiti, Interaction of hydroxyapatite and chitosan with gentamicin and their antimicrobial activities: DFT and molecular docking approach , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 3, August 2025
- I. E. Otuokere, J. G. Ohwimu, K. C. Amadi, C. O. Alisa, F. C. Nwadire, O. U. Igwe, A. A. Okoyeagu, C. M. Ngwu, Synthesis, Characterization and Molecular Docking Studies of Mn (II) Complex of Sulfathiazole , Journal of the Nigerian Society of Physical Sciences: Volume 1, Issue 3, August 2019
- Chidi Duru, Ijeoma Duru, Chiagoziem Chidiebere, Virtual Screening of Selected Natural Products as Human Tyrosinase-Related Protein 1 Blockers , Journal of the Nigerian Society of Physical Sciences: Volume 3, Issue 3, August 2021
- U. B. Amadi, M. Ogwuegbu, C. K. Enenebeaku, G. onyedika, Design, synthesis and studies on thermodynamic and biological activities of lanthanide III complexes of hydrazinecarbothioamide ligands , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 2, May 2025
- Segun Oladipo, Adesola A. Adeleke, Abosede A. Badeji, Katherine I. Babalola, Ayomide H. Labulo, Ibrahim Hassan, Sadiq T. Yussuf, Samuel O. Olalekan, Computational investigation and biological activity of selected Schiff bases , Journal of the Nigerian Society of Physical Sciences: Volume 6, Issue 3, August 2024
- M. E. Khan, C. E. Elum, A. O. Ijeomah, P. J. Ameji, I. G. Osigbemhe, E. E. Etim, J. V. Anyam, A. Abel, C. T. Agber, Isolation, Characterization, Antimicrobial and Theoretical Investigation of Some Bioactive Compounds Obtained from the Bulbs of Calotropisprocera , Journal of the Nigerian Society of Physical Sciences: Volume 5, Issue 3, August 2023
- C. E. Duru, C. E. Enyoh, I. A. Duru, M. C. Enedoh, Degradation of PET Nanoplastic Oligomers at the Novel PHL7 Target:Insights from Molecular Docking and Machine Learning , Journal of the Nigerian Society of Physical Sciences: Volume 5, Issue 1, February 2023
- K. O. Eberendu , J. I. Iheanyichukwu, O. M. Mac-kalunta, C. I. Nwankwo, I. E. Otuokere, J. C. Nnaji, Zn(II) and Fe(II) complexes of 2,4-dinitro-N-[(Z)-[(E)-3-(2-nitrophenyl)prop-2-enylidene] amino] aniline: synthesis, characterization and In Silico SARS-CoV-2 inhibition studies , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 1, February 2025
- E. E. Etim, Benchmark Studies on the Isomerization Enthalpies for Interstellar Molecular Species , Journal of the Nigerian Society of Physical Sciences: Volume 5, Issue 2, May 2023
You may also start an advanced similarity search for this article.






