Efficient-ViT B0Net: A high-performance light weight transformer for rice leaf disease recognition and classification
Keywords:
Plant disease, Deep learning, Efficient net B0, Vision transformerAbstract
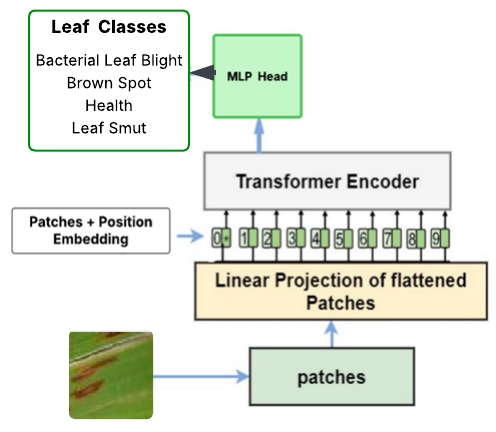
Plant disease detection has become a demanding and challenging task in today’s environment because many different types of plants exist world-wide, and very varied infections are found in them. The proposed work introduced a hybrid architecture to perform plant disease recognition and classification accurately and efficiently. The proposed model utilizes the strengths of CNN and Vision Transformer, where CNN successfully extracts local fine-grained texture features quickly. At the same time, ViT plays a vital role in extracting global and deep features from the leaf images. The suggested model was evaluated on a rice leaf dataset for paddy disease recognition and classification. The dataset consists of images representing four different types of rice leaves, with each class containing 4,000 samples. It includes healthy and diseased leaves, where the diseased category is further divided into three specific classes: Brown Spot, Bacterial Leaf Blight, and Leaf Smut. The suggested model worked well on the input dataset and achieved a testing accuracy of 99.13%. The Precision, recall, and F1 score of the proposed model were recorded as 99.13%, 99.13%, and 99.13%, respectively. The proposed method achieves a classification accuracy of 99.13%, outperforming SOTA models such as ViT-small, DenseNet121, ResNet50, EfficientNet B0 and SqueezeNet by 2–9% on the same dataset. The proposed method was compared with other approaches on the same experimental environment. These results demonstrate the effectiveness of our EfficientNet-ViT-based pipeline in capturing both local and global features for accurate rice disease classification.
Published
How to Cite
Issue
Section
Copyright (c) 2025 Santosh Kumar Upadhyay, Rajesh Prasad

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Similar Articles
- S. I. Ele, U. R. Alo, H. F. Nweke, A. H. Okemiri, E. O. Uche-Nwachi, Deep convolutional neural network (DCNN)-based model for pneumonia detection using chest x-ray images , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 2, May 2025
- Shaymaa Mohammed Ahmed, Majid Khan Majahar Ali, Raja Aqib Shamim, Integrating robust feature selection with deep learning for ultra-high-dimensional survival analysis in renal cell carcinoma , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 4, November 2025
- P. O. Odion, M. N. Musa, S. U. Shuaibu, Age Prediction from Sclera Images using Deep Learning , Journal of the Nigerian Society of Physical Sciences: Volume 4, Issue 3, August 2022
- Nahid Salma, Majid Khan Majahar Ali, Raja Aqib Shamim, Machine learning-based feature selection for ultra-high-dimensional survival data: a computational approach , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 3, August 2025
- Sherifdeen O. Bolarinwa, Eli Danladi, Andrew Ichoja, Muhammad Y. Onimisia, Christopher U. Achem, Synergistic Study of Reduced Graphene Oxide as Interfacial Buffer Layer in HTL-free Perovskite Solar Cells with Carbon Electrode , Journal of the Nigerian Society of Physical Sciences: Volume 4, Issue 3, August 2022
- V Umarani, A Julian, J Deepa, Sentiment Analysis using various Machine Learning and Deep Learning Techniques , Journal of the Nigerian Society of Physical Sciences: Volume 3, Issue 4, November 2021
- Raphael Ozighor Enihe, Rajesh Prasad, Francisca Nonyelum Ogwueleka, Fatimah Binta Abdullahi, The effect of imbalance data mitigation techniques on cardiovascular disease prediction , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 2, May 2025
- Muhammad Dahiru Liman, Salamatu Ibrahim Osanga, Esther Samuel Alu, Sa'adu Zakariya, Regularization Effects in Deep Learning Architecture , Journal of the Nigerian Society of Physical Sciences: Volume 6, Issue 2, May 2024
- Paavithashnee Ravi Kumar, Majid Khan Majahar Ali, Olayemi Joshua Ibidoja, Identifying heterogeneity for increasing the prediction accuracy of machine learning models , Journal of the Nigerian Society of Physical Sciences: Volume 6, Issue 3, August 2024
- A. E. Ibor, D. O. Egete, A. O. Otiko, D. U. Ashishie, Detecting network intrusions in cyber-physical systems using deep autoencoder-based dimensionality reduction approach anddeep neural networks , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 3, August 2025
You may also start an advanced similarity search for this article.






