A novel multi-class classification method for arrhythmias using Hankel dynamic mode decomposition and long short-term memory networks
Keywords:
Dynamic Mode Decomposition (DMD), Long Short-Term Memory (LSTM), Arrhythmia, Multi-ClassAbstract
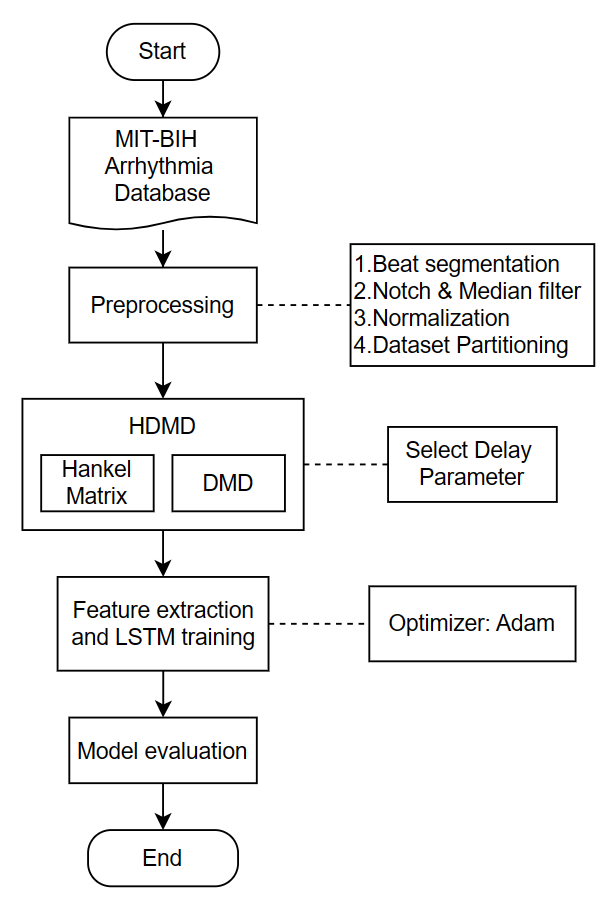
The complex dynamic properties of ECG signals and the challenge of multi-category classification make automated diagnosis of arrhythmias difficult. In this paper, we propose a new model for the multiclassification task of arrhythmia, which combines Hankel Dynamic Modal Decomposition (HDMD) and Long Short-Term Memory Network (LSTM).HDMD is used to construct the Hankel matrix, the optimal delay parameter is selected based on 90% of the energy of the singular values, and dynamic modal features extracted from it are used as the input sequences of LSTM. The LSTM model is optimisation is performed by minimising the cross-entropy loss function, setting the maximum number of iterations to 60 and using an early stopping strategy to avoid overfitting. The model was validated on the MIT-BIH arrhythmia database, which contains 109,402 beats and is classified into five categories according to the AAMI criteria: normal beats (N), ventricular premature beats (V), fusion beats (F), atrial premature beats (S) and unclassifiable (Q). By comparing with the direct use of LSTM, the experimental results showed that the HDMD[1]LSTM model showed different degrees of improvement in all five classifications, especially in the classification of atrial premature beats (S) and ventricular premature beats (V), and the overall classification accuracy improved to 0.85. Future work can focus on two aspects: first, to solve the category imbalance problem, by over sampling or undersampling techniques to improve the classification ability of a few categories; second, exploring the embedding of DMD into the network structure of LSTM to further optimise the feature extraction and classification performance.
Published
How to Cite
Issue
Section
Copyright (c) 2025 Chuchu Liang, Majid Khan Majahar Ali, Lili Wu

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Most read articles by the same author(s)
- O. J. Ibidoja, F. P. Shan, Mukhtar, J. Sulaiman, M. K. M. Ali, Robust M-estimators and Machine Learning Algorithms for Improving the Predictive Accuracy of Seaweed Contaminated Big Data , Journal of the Nigerian Society of Physical Sciences: Volume 5, Issue 1, February 2023
- Xiaojie Zhou, Majid Khan Majahar Ali, Farah Aini Abdullah, Lili Wu, Ying Tian, Tao Li, Kaihui Li, Air quality prediction enhanced by a CNN-LSTM-Attention model optimized with an advanced dung beetle algorithm , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 3, August 2025
- Shaymaa Mohammed Ahmed, Majid Khan Majahar Ali, Raja Aqib Shamim, Integrating robust feature selection with deep learning for ultra-high-dimensional survival analysis in renal cell carcinoma , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 4, November 2025
- Paavithashnee Ravi Kumar, Majid Khan Majahar Ali, Olayemi Joshua Ibidoja, Identifying heterogeneity for increasing the prediction accuracy of machine learning models , Journal of the Nigerian Society of Physical Sciences: Volume 6, Issue 3, August 2024
- Raja Aqib Shamim, Majid Khan Majahar Ali, Optimizing discrete dutch auctions with time considerations: a strategic approach for lognormal valuation distributions , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 1, February 2025
- Shaymaa Mohammed Ahmed, Majid Khan Majahar Ali, Arshad Hameed Hasan, Evaluating feature selection methods in a hybrid Weibull Freund-Cox proportional hazards model for renal cell carcinoma , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 3, August 2025
- Nahid Salma, Majid Khan Majahar Ali, Raja Aqib Shamim, Machine learning-based feature selection for ultra-high-dimensional survival data: a computational approach , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 3, August 2025
- xiaojie zhou, Majid Khan Majahar Ali, Farah Aini Abdullah, Lili Wu, Ying Tian, Tao Li, Kaihui Li, Implementing a dung beetle optimization algorithm enhanced with multi-strategy fusion techniques , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 2, May 2025
- Ibrahim Adamu Mohammed, Majid Khan Majahar Ali, Sani Rabiu, Raja Aqib Shamim, Shahida Shahnawaz, Development and validation of hybrid drying kinetics models with finite element method integration for black paper in a v-groove solar dryer , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 4, November 2025
- Raja Aqib Shamim, Majid Khan Majahar Ali, Mohamed Farouk Haashir bin Hamdullah, Computational optimization of auctioneer revenue in modified discrete Dutch auctions with cara risk preferences , Journal of the Nigerian Society of Physical Sciences: Volume 8, Issue 1, February 2026 (In Progress)