Integrating robust feature selection with deep learning for ultra-high-dimensional survival analysis in renal cell carcinoma
Keywords:
Ultra-High-Dimensional Survival Analysis, Renal Cell Carcinoma (RCC), Feature Selection with Deep Learning, Robust SIS, Robust ISISAbstract
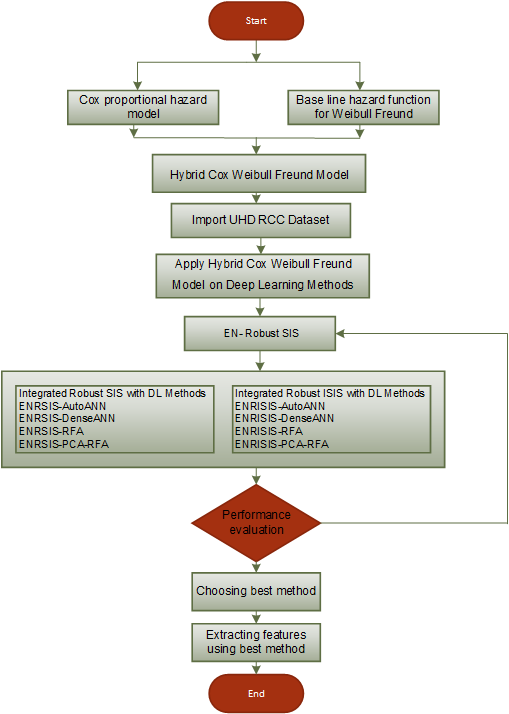
The research method applies robust feature selection approaches to ultra-high-dimensional survival data records from Renal Cell Carcinoma patients through deep learning methodologies. The linear methods LASSO and Elastic Net encounter failure when processing data because they face simultaneous multicollinearity issues in addition to overfitting effects and produce marginal survival outcome variability prediction at 54%. We suggest combining ISIS with deep learning architectures featuring PCA-RFA-RSIS models as a remedy to handle these present limitations. Among all evaluated methods PCA-RFA-RSIS is proved most accurate with an MSE measurement of 24.39 and R2 value of 0.89. PCA improved the model’s dimensionality reduction power and robust ISIS maintained model stability despite outliers present in the data. The discovery holds significant value in precision medicine because it creates opportunities to develop individualized therapy for kidney failure patients. Further research needs to enhance hybrid models and expand their utilization between different diseases as well as complex biological systems.
Published
How to Cite
Issue
Section
Copyright (c) 2025 Shaymaa Mohammed Ahmed, Majid Khan Majahar Ali, Raja Aqib Shamim

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Most read articles by the same author(s)
- O. J. Ibidoja, F. P. Shan, Mukhtar, J. Sulaiman, M. K. M. Ali, Robust M-estimators and Machine Learning Algorithms for Improving the Predictive Accuracy of Seaweed Contaminated Big Data , Journal of the Nigerian Society of Physical Sciences: Volume 5, Issue 1, February 2023
- Xiaojie Zhou, Majid Khan Majahar Ali, Farah Aini Abdullah, Lili Wu, Ying Tian, Tao Li, Kaihui Li, Air quality prediction enhanced by a CNN-LSTM-Attention model optimized with an advanced dung beetle algorithm , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 3, August 2025
- Paavithashnee Ravi Kumar, Majid Khan Majahar Ali, Olayemi Joshua Ibidoja, Identifying heterogeneity for increasing the prediction accuracy of machine learning models , Journal of the Nigerian Society of Physical Sciences: Volume 6, Issue 3, August 2024
- Raja Aqib Shamim, Majid Khan Majahar Ali, Optimizing discrete dutch auctions with time considerations: a strategic approach for lognormal valuation distributions , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 1, February 2025
- Shaymaa Mohammed Ahmed, Majid Khan Majahar Ali, Arshad Hameed Hasan, Evaluating feature selection methods in a hybrid Weibull Freund-Cox proportional hazards model for renal cell carcinoma , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 3, August 2025
- Nahid Salma, Majid Khan Majahar Ali, Raja Aqib Shamim, Machine learning-based feature selection for ultra-high-dimensional survival data: a computational approach , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 3, August 2025
- Chuchu Liang, Majid Khan Majahar Ali, Lili Wu, A novel multi-class classification method for arrhythmias using Hankel dynamic mode decomposition and long short-term memory networks , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 2, May 2025
- xiaojie zhou, Majid Khan Majahar Ali, Farah Aini Abdullah, Lili Wu, Ying Tian, Tao Li, Kaihui Li, Implementing a dung beetle optimization algorithm enhanced with multi-strategy fusion techniques , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 2, May 2025
- Ibrahim Adamu Mohammed, Majid Khan Majahar Ali, Sani Rabiu, Raja Aqib Shamim, Shahida Shahnawaz, Development and validation of hybrid drying kinetics models with finite element method integration for black paper in a v-groove solar dryer , Journal of the Nigerian Society of Physical Sciences: Volume 7, Issue 4, November 2025
- Raja Aqib Shamim, Majid Khan Majahar Ali, Mohamed Farouk Haashir bin Hamdullah, Computational optimization of auctioneer revenue in modified discrete Dutch auctions with cara risk preferences , Journal of the Nigerian Society of Physical Sciences: Volume 8, Issue 1, February 2026 (In Progress)